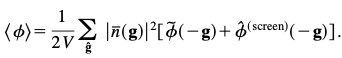Solution LAB2 guanine 2
- Back to the previous page: Electronic properties of isolated molecules#Exercise2
Step 1
To calculate HOMO and LUMO level perform an scf and next an nscf calculation adding empty states (e.g. nbnd = 36) Then you can inspect the eigenvalues or:
> grep "highest occupied, lowest unoccupied level"
Step 2
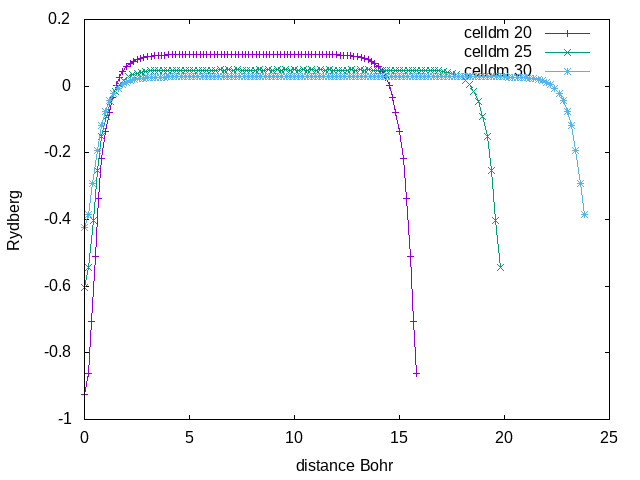
Repeat the same calculation of step 1 by changing the volume of your supercell (e.g celldm(1)=20,25,30). You can also use a script to automatise the runs.
Then collect the results of the energy level as a function of the volume. You can observe a strong dependency of the eigenvalues from the volume of the supercell.
Step 3
Step 4
We can isolate the system in the supercell using an algorithm to correctly treat long-range interactions. In the &SYSTEM name-list insert:
assume_isolated='mt'
This is the Martyna-Tuckerman method: A reciprocal space based method for treating long range interactions in ab initio and force-field-based calculations in clusters G. Martyna, M. Tuckerman J. Chem. Phys. 110 2810 (1999)
The new function appearing in the average cluster potential energy, is the difference between the Fourier series and Fourier transform form of the long range part of the potential energy evaluated at the quantized g-vector and will be referred to as the screening function because it is constructed to ‘‘screen’’ the interaction of the system with an infinite array of periodic images.