Difference between revisions of "Solution LAB2 guanine"
(→Step 2) |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | * Back to the previous page: [[Electronic properties of isolated molecules# | + | * Back to the previous page: [[Electronic properties of isolated molecules#Exercise 1]] |
| − | ==Step 1== | + | ==Step 1: geometry building== |
| − | *Once you have Avogadro installed, open it and let's use the molecular builder to visualise a Guanine molecule: | + | * Once you have Avogadro installed, open it and let's use the molecular builder to visualise a Guanine molecule: |
| − | Build --> Insert-->Fragment-->nucleobases-->guanine | + | Build --> Insert-->Fragment-->nucleobases-->guanine |
Click on the selection settings "arrow" and right click to visualise the molecule. | Click on the selection settings "arrow" and right click to visualise the molecule. | ||
| Line 10: | Line 10: | ||
Clicking on manipulate settings "hand" you can then rotate the molecule. | Clicking on manipulate settings "hand" you can then rotate the molecule. | ||
| − | *Now we perform a classical relaxation using an Avogadro built-in Force Filed | + | * Now we perform a classical relaxation using an Avogadro built-in Force Filed |
| − | Extension-->Molecular Mechanics and we choose a FF | + | Extension-->Molecular Mechanics and we choose a FF |
| + | Extension-->Optimize geometry | ||
| − | + | In few steps we have our relaxed geometry that we can save in .xyz format using: | |
| − | + | File-->Save as-->select .xyz format and save in your disk as guanine_FF.xyz | |
| + | |||
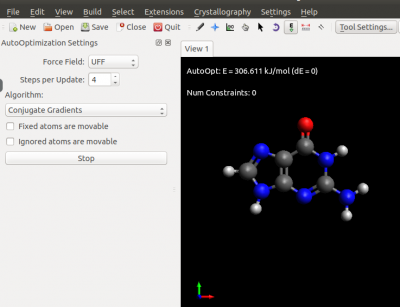
| + | The result of your FF optimisation should look like the following | ||
| − | File | + | [[File:Snapshot avogadro.png|none|400px|top|]] |
==Step 2== | ==Step 2== | ||
| − | Now we are ready to perform a | + | Now we are ready to perform a DFT simulations. As a first step, update your pseudo potentials directory by using git commands: |
git pull | git pull | ||
| Line 27: | Line 30: | ||
new pseudo potentials (Nitrogen and Oxygen) will be downloaded in your pseudo potential directory. | new pseudo potentials (Nitrogen and Oxygen) will be downloaded in your pseudo potential directory. | ||
| − | Create a | + | Create a scf input file (guanine_scf.in) paying attention to: |
| − | + | ||
ibrav=8 #orthorombic cell with vacuum | ibrav=8 #orthorombic cell with vacuum | ||
celldm(1) = 25.00 | celldm(1) = 25.00 | ||
| Line 46: | Line 49: | ||
ATOMIC_POSITIONS (angstrom) | ATOMIC_POSITIONS (angstrom) | ||
| − | '''Important''': How many k points you need? | + | '''Important''': |
| + | * How many '''k'''-points you need? | ||
| + | * the amount of vacuum needs to be converged | ||
| + | * same for the kinetic energy cutoff (note that here we can use the value of the total force as a convergence parameter) | ||
| + | |||
| + | # ecutwfc Ry | ||
| + | 40 Total force = 0.316709 Total SCF correction = 0.000067 | ||
| + | 50 Total force = 0.315474 Total SCF correction = 0.000089 | ||
| + | 60 Total force = 0.319550 Total SCF correction = 0.000100 | ||
| + | 80 Total force = 0.322261 Total SCF correction = 0.000098 | ||
| + | 100 Total force = 0.323131 Total SCF correction = 0.000119 | ||
| + | 120 Total force = 0.323153 Total SCF correction = 0.000140 | ||
| + | |||
| + | Despite 60-80 Ry seem to be appropriate, here we decide to use <code>ecutwfc=50</code> in order to keep the calculation time under control. | ||
| + | |||
| + | |||
| + | ==Step 3== | ||
| + | We can now run a QM relaxation using DFT. | ||
| + | Edit the input file according to: | ||
| + | &CONTROL | ||
| + | [...] | ||
| + | calculation="relax" | ||
| + | etot_conv_thr=1.0D-4 | ||
| + | forc_conv_thr=1.0D-3 | ||
| + | # we set a medium-tight threshold in order not to have a too long calculation | ||
| + | / | ||
| + | and add the <code>&IONS</code> namelist. | ||
| + | |||
| + | Run the relaxation, it will take a while to rearrange the atoms in the relaxed positions according to the threshold parameter. | ||
| + | |||
| + | $> pw.x < guanine_relax.in > guanine_relax.out & | ||
| + | |||
| + | One the job has converged you can compare the initial position (Force Field relaxed) with the final ones (QM relaxed). You can use xcrysden to visualise the optimisation steps. | ||
| + | |||
| + | xcrysden --pwo guanine_relax.out | ||
| + | |||
| + | you can reduce the dimensionality to 0D and choose the option: Display All Coordinates as Animation | ||
| + | In this way you will have a movie of the molecule relaxation | ||
Latest revision as of 11:28, 26 March 2021
- Back to the previous page: Electronic properties of isolated molecules#Exercise 1
Step 1: geometry building
- Once you have Avogadro installed, open it and let's use the molecular builder to visualise a Guanine molecule:
Build --> Insert-->Fragment-->nucleobases-->guanine
Click on the selection settings "arrow" and right click to visualise the molecule.
Clicking on manipulate settings "hand" you can then rotate the molecule.
- Now we perform a classical relaxation using an Avogadro built-in Force Filed
Extension-->Molecular Mechanics and we choose a FF Extension-->Optimize geometry
In few steps we have our relaxed geometry that we can save in .xyz format using:
File-->Save as-->select .xyz format and save in your disk as guanine_FF.xyz
The result of your FF optimisation should look like the following
Step 2
Now we are ready to perform a DFT simulations. As a first step, update your pseudo potentials directory by using git commands:
git pull
new pseudo potentials (Nitrogen and Oxygen) will be downloaded in your pseudo potential directory.
Create a scf input file (guanine_scf.in) paying attention to:
ibrav=8 #orthorombic cell with vacuum celldm(1) = 25.00 celldm(2) = 1.0 celldm(3) = 0.8
Set the correct number and types of atoms. Set all the atomic species using the psuedo you have downloaded:
H 1.0 H.pz-vbc.UPF C 1.0 C.pz-vbc.UPF N 1.0 N.pz-vbc.UPF O 1.0 O.pz-mt.UPF
insert the atomic positions in Angstrom:
ATOMIC_POSITIONS (angstrom)
Important:
- How many k-points you need?
- the amount of vacuum needs to be converged
- same for the kinetic energy cutoff (note that here we can use the value of the total force as a convergence parameter)
# ecutwfc Ry 40 Total force = 0.316709 Total SCF correction = 0.000067 50 Total force = 0.315474 Total SCF correction = 0.000089 60 Total force = 0.319550 Total SCF correction = 0.000100 80 Total force = 0.322261 Total SCF correction = 0.000098 100 Total force = 0.323131 Total SCF correction = 0.000119 120 Total force = 0.323153 Total SCF correction = 0.000140
Despite 60-80 Ry seem to be appropriate, here we decide to use ecutwfc=50 in order to keep the calculation time under control.
Step 3
We can now run a QM relaxation using DFT. Edit the input file according to:
&CONTROL [...] calculation="relax" etot_conv_thr=1.0D-4 forc_conv_thr=1.0D-3 # we set a medium-tight threshold in order not to have a too long calculation /
and add the &IONS namelist.
Run the relaxation, it will take a while to rearrange the atoms in the relaxed positions according to the threshold parameter.
$> pw.x < guanine_relax.in > guanine_relax.out &
One the job has converged you can compare the initial position (Force Field relaxed) with the final ones (QM relaxed). You can use xcrysden to visualise the optimisation steps.
xcrysden --pwo guanine_relax.out
you can reduce the dimensionality to 0D and choose the option: Display All Coordinates as Animation In this way you will have a movie of the molecule relaxation